 |
|||||||||||||||
Our laboratory studies the major human fungal pathogen Candida albicans. Although present as a commensal in the digestive tract of most healthy people, C. albicans is also capable of causing a wide variety of systemic and mucosal infections. Immunocompromised individuals such as AIDS patients, organ transplant recipients, cancer patients undergoing chemotherapy and recipients of artificial joints and prosthetic devices are particularly susceptible to infection. C. albicans is also a highly evolved fungal pathogen, capable of invading nearly every organ and tissue in the human body; indeed, there are no known reservoirs for C. albicans outside the mammalian host.
Figure 1. The blastospore to filament transition. The C. albicans blastospore to filament transition can occur in response to a variety of host environmental signals. Filaments occur in pseudohyphal (with constrictions at the septa) or true hyphal form. Photographs indicate colony morphologies of cells in the blastospore or filamentous forms. How is C. albicans able to function so effectively as a pathogen? This organism possesses a number of properties which contribute to its virulence, including secretion of degradative enzymes and adhesion to host cells. Research in our laboratory focuses on one such virulence property, the ability to undergo a
reversible morphological transition from blastospores (single round cells) to filaments (elongated cells attached end-to-end, see Figure 1). Previous studies have shown that this morphological transition is required for virulence and can occur in response to a wide variety of host inducing signals. In order to gain a better understanding of the mechanisms that control the C. albicans morphological transition in response to specific host environmental cues we have taken a genomic approach. Using whole-genome C. albicans DNA microarrays (Figure 2) we have identified a set of 61 genes that are induced during the blastospore to filament transition in response to serum and body temperature (37°C). We are specifically interested in determining: 1) the transcriptional mechanisms that control induction of the C. albicans filamentous growth program in response to specific host inducing signals, 2) the mechanisms by which individual genes in the C. albicans filamentous growth program function to establish and maintain infection in host tissues:
Figure 2. C. albicans DNA microarray. Each spot represents DNA from a single gene in the C. albicans genome. cDNAs prepared from strains of interest are coupled to fluorescently labelled dyes and hybridized to the array. Color differences between individual spots indicate changes in gene expression. Transcriptional Regulators of C. albicans Filamentous Growth and Virulence
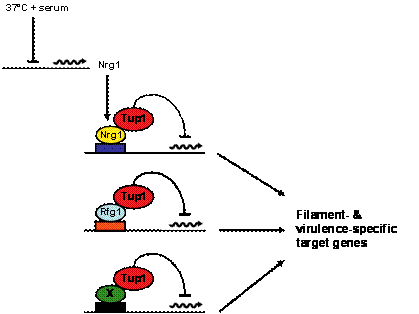
We have previously identified and characterized three key transcriptional repressors of the C. albicans blastospore to filament transition. Two of these regulators, Rfg1 and Nrg1, function as promoter-specific DNA-binding proteins and are known to direct repression by recruitment of the third protein, Tup1 corepressor, to the promoters of filament- and virulence-specific target genes. Using comparative DNA microarray analysis we have determined that approximately one-half of all genes in the C. albicans filamentous growth program are under negative control by Rfg1, Nrg1 and/or Tup1. We (and others) have also shown that the NRG1 transcript is down-regulated in response to serum and 37°C, suggesting a model whereby induction of the C. albicans filamentous growth program occurs by the relief of transcriptional repression (Figure 3). Current research is focused on the mechanisms that control the expression of transcriptional regulators of filamentous growth in response to specific host inducing conditions. A variety of genetic, genomic, molecular and biochemical approaches are being taken to address this question.
Figure 3. Model for control of C. albicans filamentous growth and virulence. Under non-filament-inducing conditions Rfg1, Nrg1 and at least one additional DNA-binding protein (X) bind to filament- and virulence-specific target gene promoters and recruit the Tup1 corepressor. In the presence of filament-inducing conditions, repression is relieved; in the case of Nrg1-Tup1 repression this occurs by down-regulation of the NRG1 transcript (adapted from Kadosh & Johnson, Mol. Biol. Cell, 2005). Identification and Characterization of Novel C. albicans Virulence Factors
The C. albicans filamentous growth program is comprised of genes involved in a wide variety of biological processes, several of which had not previously been implicated in filamentous growth and virulence. We have identified four gene classes which appear to be significantly over-represented in the filamentous growth program compared to their representation in the genome as a whole: 1) cell wall components, 2) cell division genes, 3) secreted/degradative enzymes, 4) ER to Golgi transport and secretion genes. The C. albicans filamentous growth program also contains a significant number of genes of unknown function. Several of these genes are unique to C. albicans, suggesting the possibility that they may be important for novel virulence mechanisms. Efforts are currently underway to further characterize these genes and determine the precise roles that they play in allowing C. albicans to establish and maintain infection in host tissues.
Publications:
- DeRubertis, F., Kadosh, D., Henchoz, S., Pauli, D., Reuter, G., Struhl, K. and Spierer, P. (1996). The histone deacetylase Rpd3 counteracts genomic silencing in Drosophila and yeast. Nature 384: 589-591.
- Kadosh, D. and Struhl K. (1997). Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365-371.
- Kadosh, D. and Struhl K. (1998). Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12: 797-805.
- Kadosh, D. and Struhl K. (1998). Targeted the recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18: 5121-5127.
- Struhl, K., Kadosh, D., Keaveney, M., Kuras, L. and Moqtaderi, Z. (1998). Activation and repression mechanisms in yeast. Cold Spring Harbor Symposium Quant. Biol. 63: 413-421.
- Kadosh, D. and Johnson, A.D. (2001). Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21: 2496-2505
- *Braun, B.R., *Kadosh, D. and Johnson, A.D. (2001). NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament-induction. EMBO J. 20: 4753-4761.
- Kadosh, D. and Johnson, A.D. (2005). Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis: Mol. Biol. Cell 16: 2903-2912.
* These authors made equal contributions.
 |
Links provided from Health Science Center pages to other web sites do not constitute or imply an endorsement of those sites, their content, or products and services associated with those sites. |